ISU INBRE Mentors

Interested in the INBRE Fellows Program?
Students - Browse the Mentor profiles below and at the Idaho INBRE official website to find potential research projects. Contact your chosen Mentor before submitting your application to be sure they are able to mentor you if you are selected as a Fellow. A letter of support from your chosen Mentor is a required component of the ISU INBRE application.
Faculty - Want to become an ISU-INBRE Mentor? Find out more under the opportunities section.

Devaleena Pradhan
Associate Professor
Summary: My research goal is to understand the physiological mechanisms of behavioral plasticity in organisms. I study a sexually plastic marine fish, the bluebanded goby, that displays adult sex change. Its sex is determined by the social group it lives in and can transform from male to female or from female to male. In our lab, we use an integrative approach that includes molecular, subcellular, and systemic levels of biological organization. We investigate endocrinological mechanisms by which neural, sensory and motor systems adapt to a changing external environment. We use in vitro and in vivo biochemical manipulations, molecular assays, hormone measurements, in-depth behavioral analyses, morphometrics, and measures of reproductive success to assess the hormonal regulation (in both long-term and short-term) of reproductive phenotypes of free-living and captive species fish. While I am trained as a neuroendocrinologist and most of my work involves neural mechanisms in the context of regulation of social behavior, I have developed interests in other local signaling mechanisms in specific tissues (e.g. reproductive tissues, muscle, spinal cord) that either undergo dramatic changes associated with life history or functional specialization. I plan to further my research in studying how the social or physical environment can regulate neural, muscle, and reproductive physiology in males and females.
Minimum classes: Physiology
Projects:
- Role of brain hormones in regulating social behavior and sex change in a marine fish. This project will entail analyzing changes in dominance behavior in fish living in small social groups. You will also learn how to homogenize tissue samples, extract hormones using solid phase extraction technique and detect hormones using enzymeimmunoassays.
- mRNA expression of steroid synthesizing enzymes and growth factors in muscles during parenting. Male gobies care for developing eggs by fanning and rubbing them until hatch. These behaviors entail intense whole-body movement including their pectoral, dorsal and caudal fins. Males also have higher levels of the androgen, ketotestosterone, in the muscles attached to the dorsal fin compared to females. In this project, you will learn to use molecular biology approaches such as RNA extraction, cDNA synthesis and PCR.
- Histological analysis of gonads and brain during sex change. Sex change involves gonadal rearrangement and rewiring of the nervous system during that process. In this project, you will learn to prepare slices of tissues using a cryostat and then using immunocytochemistry to identify enzymes, hormones, and receptors that are localized in different regions of the gonad and brain.

Heather Ray
Assistant Professor
Summary: Embryonic development is a highly intricate and complex process that allows a fertilized egg to generate all of the cells and tissues of a living multicellular organism. These cellular processes are driven by gene expression that is tightly regulated in both time (different steps during development) and space (different places within the embryo). Mutations in genes that are important for embryonic development can disrupt these important processes and lead to developmental disorders such as spina bifida and cleft lip and palate which are quite common in the human population. In order to figure out ways to prevent such disorders from occurring, we need to understand both how normal development happens, and how disrupting normal development leads to defects. In my lab, we have identified several genes that, when mutated, result in human developmental disorders. Our work aims to understand how each of these genes function in embryonic development and how mutation in each of these genes would actually disrupt that function to lead to a developmental disorder. To address these questions, we use the African Clawed frog as they share both genetic and cellular similarity to humans in these early developmental processes. After performing in vitro fertilization, frog embryos will develop in the lab and we can study these processes in real time using a combination of molecular biology, microscopy, cell biology and classic embryology techniques.
Minimum classes: Cell biology, genetics
Projects:
Loss of function of the gene masp1/3 disrupts embryonic development
Mutations in masp1/3 have been identified in human patients with the developmental disorder 3MC Syndrome. In this project, we will decrease the expression of masp1/3 in frog embryos and determine how that affects development. This project will involve careful observation and microscopy, reverse transcription PCR, and in situ hybridization techniques
The gene hic1 regulates Wnt signaling during neural crest development
Mutations in the gene hic1 disrupt development of a population of cells called the neural crest. In this project we will look more closely at how hic1 functions by regulating an important signaling pathway called Wnt that directs these cells in their development. This project will involve beginning bioinformatics, PCR and plasmid-based cloning, and in situ hybridization

James Groome
Professor - Neuroscience
Office: Office: LS 136B
Summary: Our laboratory investigates the molecular basis of ion channel function. These proteins include the voltage-gated channels of nerve and muscle and the neurotransmitter receptors in the brain and periphery. We use a diversity of approaches to study these channels and the defects caused by mutations associated with disease states. Molecular biology techniques include site directed mutagenesis to reiterate the mutations in channels found in patients with channelopathy diseases such as myotonia and periodic paralysis, and microbiology techniques to work with the recombinant DNA with the goal of expressing these proteins in the oocytes of Xenopus frogs. These oocytes serve as the expression system for our electrophysiological approach to study ion channels. Two electrode voltage clamp experiments are employed to study the response to acetylcholine for nicotinic acetylcholine receptors that are an important determinant of dopamine release and thus a target in Parkinson’s Disease therapy. Our third approach is computational. We use sequence alignments and homology algorithms to create three dimensional models of the membrane proteins mentioned above and study their function in silico. We also use mathematical models of excitable cell membranes such as skeletal muscle fibers to investigate the functional impact of the channel defects determined with electrophysiology on action potential signaling to study the basis of paramyotonia congenita and other channelopathies of muscle fibers.
Minimum Classes: N/A

Jason Pilarski
Associate Professor
Office: Life Sciences 136A
Summary: My laboratory is interested in how individual neurons and neural circuits produce maintain and modulate spontaneous rhythmic electrical activity in the brain and how this early activity is transformed into functional neural circuits. Spontaneous rhythms are important central nervous system phenomena that underlie repetitive motor behaviors such as locomotion, chewing, and breathing to name only a few. The mechanisms involved in this type of neural activity include the actions of individual cells and also the nature of cell-to-cell connectivity that can involve many hundreds of neurons. We study these neural networks by exploring neural activity, anatomy, synaptic neurophysiology, biochemistry and pharmacology using a live brain preparations from avian embryos. This brain preparation produces spontaneous neural activity even when isolated from the rest of the nervous system. Specifically, my laboratory is interested in brainstem circuits that underlie the ability to regulate and control breathing patterns. Breathing is both an amazingly regular and consistent, repetitive motor behavior (i.e., during an 80 year lifespan we can produce billions of breathing cycles), yet breathing is also a behavior that is highly modulated and plastic. For example, breathing is altered in a breath-by-breath manner during speaking, swallowing, coughing, and exercise. The breathing circuit is also modified during development, and my laboratory’s main focus is to understand both the normal and pathological development of breathing circuits during early life. Recently, we showed that brainstem breathing circuits are susceptible to pathological alterations during critical periods of development, which may help explain some medical problems experienced by newborns, such as obstructive apnea and sudden infant death syndrome.
Minimum classes: Any introductory science classes; biology, physics, chemistry preferred
Projects:
1. To isolate the specific neural circuit in the brainstem that is responsible for spontaneous breathing in birds. This circuit is completely undescribed. It is important because birds are vertebrate animals that breath just like us, yet they develop in a way (via the external egg) that allows for manipulation and experimentation that is nearly impossible in humans or even other mammals. This would be the first step in developing a model of developmental neural plasticity that is critical for understanding how neural circuits change in early life, especially when subject to altered neurochemical environments. For this work, we employ electrical recordings from live neurons and entire neural circuits using glass electrodes, amplifiers, and audiovisual equipment. This work also involves microsurgical skills, anatomy, statistics and understanding of the central nervous system environments.
2. To understand the neurochemical basis for breathing rhythm(s) in birds. In mammals, the basic circuit is unique compared to the rest of the animal kingdom and also not well understood. If we could understand the basic neurochemical environment in other “simpler” animals we could understand ourselves better. For these experiments, electrical recording equipment will be used (see also project #1) and pharmacological concepts will also be employed. The student will gain a thorough understanding of chemical communication (i.e., synaptic transmission) in the central nervous system.
3. To understand the role of homeostatic plasticity in early breathing circuits. Homeostatic plasticity is the employment of mechanisms that act to stabilize the activity of a neuron or neuronal circuit around some set-point value. Homeostatic plasticity is presumably an important phenomenon that ensures stabilization of breathing rhythms for regulating the internal electrochemical environment. However, little is known about these mechanisms during early development when the embryo and it’s environment is rapidly changing. The student will gain a thorough understanding of the brainstem and mechanisms of short and long term plasticity.

Julia Martin
Associate Professor
Office: Life Sciences 329
Summary: We are interested in how bacteria sense, respond, and adapt to metal ion changes in their surrounding environment. More specifically, we study the physiological underpinnings and molecular mechanisms of how cells regulate transition metal ion homeostasis and manganese detoxification in the model Gram-positive bacterium and respiratory pathogen Streptococcus pneumoniae.
Manganese is an essential trace nutrient that serves as a required cofactor in bacteria and is largely recognized for its role in oxidative stress resistance and in enhancing virulence of bacterial pathogens. However, when in excess, manganese can impair bacterial growth, the mechanism of which is poorly explored. Thus far, we have shown that excess manganese causes an imbalance in intracellular transition metal ion pools, which potentially affects enzyme activity. Current studies seek to (i) further our understanding of the regulation and trafficking of intracellular manganese and (ii) identify enzyme targets that are disrupted during manganese-stress in S. pneumoniae.
Minimum classes: General Biology, General Chemistry, Microbiology, Biochemistry
Projects: We use a variety of techniques and approaches that span the interface of biology and chemistry to study manganese homeostasis and detoxification. Classical microbiology and molecular biology techniques include microbial culture, microscopy, genetic manipulations, site-directed mutagenesis, molecular cloning, and quantitative real-time PCR. Biochemistry techniques include protein expression and purification, enzyme assays, protein interactions, western blot, and structure modeling.
Possible project areas include:
1) Investigating the impact of metal bioavailability on cellular growth and metabolism in pneumococcus.
2) Understanding how pneumococcal capsule polysaccharide is regulated, a key aspect in virulence.
3) Identifying and characterizing metalloprotein virulence factors that contribute to pneumococcal pathogenesis.

Kinta Serve
Associate Professor
Summary: Our lab is interested in how exposure to environmental contaminants affects the immune system. Specifically, we use asbestos exposure as a model of environment-induced autoimmunity. Asbestos is a general term applied to a group of mineral fibers. Asbestos exposure can occur through commercial use (usually to chrysotile fibers) or through exposure to natural asbestos outcroppings; the latter typically occurs when people recreate or with new construction projects that disturb the minerals. Asbestos exposures are often associated with pulmonary disease, including lung cancer, interstitial fibrosis, and mesothelioma. However, we have found that exposure to amphibole asbestos (a straight-chained fiber) is also linked to an increased incidence of pleural fibrosis, systemic autoimmune disease, and increased production of autoantibodies.
The overall goals for our lab include: 1) examining how amphibole fibers alter immune responses, 2) identify cell targets for asbestos-induced autoantibodies, and 3) test therapeutics to reduce immune responses and fibrosis following asbestos exposure.
Minimum classes:
- Cell biology or Microbiology (required)
- Immunology (optional)
- Molecular biology (optional)
- A&P (optional)
Projects:
1) Neutrophils are some of the first immune cells to respond to infection. Similarly, they are some of the first cells recruited to sites of asbestos deposition. Therefore, we are interested in understanding how asbestos fibers affect these cells and in turn how these effects may alter immune responses to fibers. The fellow working on this project will use techniques like cell culture, microscopy, and flow cytometry to determine the mechanisms of neutrophil response to asbestos fibers.
2) B cells are part of the adaptive immune system and implicated in the development of autoimmune disease through production of autoantibodies. We suspect that asbestos fiber exposure can change the function or differentiation of B cells into different subsets. This project aims to identify B cell responses to asbestos fibers and to measure B cell responses following exposures by using cell culture, flow cytometry, and animal models.
3) We have previously reported that a synthetic lignan, LGM2605, reduces early immune responses to asbestos exposure. An on-going lab project is to characterize both innate and adaptive immune responses following LGM2605 treatment and to determine if this compound can reduce asbestos-associated fibrosis and autoimmune disease. The fellow working on this project will learn histology, microscopy, ELISA, and flow cytometry techniques.

Kristin Lane
Assistant Professor
Office: Life Science 325
Summary: Multidrug resistant (MDR) pathogens are a global threat and a challenge for modern medicine. As MDR pathogens outpace the development of new therapeutics, it is critical to develop alternative treatment strategies. The Lane Lab studies Plasmodium falciparum parasites, which cause malaria in humans, and kills almost a half-million people annually, creating a desperate need for new interventions.
In the human malaria parasite, Plasmodium falciparum, mitochondrial function is critically essential in both the human and mosquito life stages. Despite being a validated drug target for decades, most mitochondrial proteins and pathways remain uncharacterized. The lack of knowledge regarding basic functions and their regulation creates an enormous gap in our ability to exploit mitochondrial biology and biochemistry for targeted drug development. It is challenging to Investigate the malaria parasite mitochondrial components. There is only a single mitochondrion per parasite for most of the life cycle. The multicopy genomes are linear or circular, encode three essential electron transport chain (ETC) genes, but no tRNAs for translation. Thus, the parasite mitochondrion has evolved noncanonical means of gene expression and regulation compared to other eukaryotes.
Minimum classes: NA
Projects: INBRE students can participate in many different aspects of ongoing projects in my laboratory, including: 1) Genetic editing of Plasmodium falciparum membrane proteins to facilitate separation of fluorescent organelles to determine the proteome of the mitochondria and apicoplast (a parasite-specific organelle). 2) Building a targeted DNA exonuclease to degrade linear DNA fragments in the parasite mitochondria 3) Analysis of the proteome dataset to identify proteins involved in gene expression of the mitochondrial genes.
Projects 1 and 2 will use standard molecular biology techniques (PCR, DNA isolation, restriction enzyme analysis, gel electrophoresis, etc) to build plasmids used for CRISPR/Cas9 editing membrane protein genes to be fluorescent. Project 3 will not be available until after summer 2023.

Peter Sheridan
Professor
Office: Garrison Hall 607
Summary: My main interests are in Microbial Diversity, the Molecular Adaptation of Enzymes to Extreme Environments, and the interplay of pathogenic microorganisms, bacteriophage, and natural antimicrobial products.
Minimum Classes: General Microbiology
Projects: Summer Fellows would be able to work on projects exploring the bacterial and fungal diversity of environmental samples; the cloning, overexpression, and characterization of genes encoding cold-active enzymes; the sequencing of the complete genomes of novel bacteria; or the genetic and metabolic response of pathogenic bacteria and bacteriophage. The student will have significant input on project choice, based on their individual interests. All of these projects rely heavily on a combination of Molecular Microbiological techniques, Biochemical techniques, and standard Microbiological techniques.

Andy Holland
Professor
Office: Office: PSC 352, Pocatello
Summary: I study the reactions of molecules that combine transition metal atoms with carbon-based groups, with particular interest in the way that the interactions between them guide the design of compounds that might be useful for catalysis or the preparation of electronic materials.
Minimum Courses: Organic Chemistry and Lab Preferred, but not strictly required
Projects: Students will prepare molecular precursors for nanomaterials using a mixture or organic and inorganic synthetic techniques. Current projects involve cerium complexes used for fundamental mechanistic studies in nanocrystal formation, and first row metals used in earth-abundant alternatives to current semiconductor technology. In both cases we are interested in making these complexes and seeing how they react. These projects sometimes require air-sensitive equipment such as an inert atmosphere glovebox. Students will also monitor related reactions and characterize products using NMR spectroscopy, thermogravimetric analysis, and other spectroscopic techniques.

Caryn Evilia
Professor
Office: PSC 356
Summary: My lab studies the structure and stability of proteins by exploring the biochemistry of the tRNA synthetases. These enzymes carry out a required cellular reaction- the attachment of the amino acid to its cognate tRNA. Without this ability, cells would die. Because of this essential function, tRNA synthetases are highly conserved in all organisms and therefore, would be an excellent target for control in pathogenic organisms. Extremophiles, organisms that live in extreme environments, have similarities to pathogens. They typically have to live in extremes in the host, in order to survive. My lab’s background on extremophiles, gives us insight into the extreme nature of how these pathogens have to live to survive, which includes how they make and structure their proteins to meet the demands of their extreme environment.
Minimum Classes: General chemistry, general biology, organic chemistry and general microbiology
Projects: Using protein alignments, we have found that a few tRNA synthetases in the pathogenic bacteria,Prevotella intermedia and Porphyromonas gingivalis, have similar features to the tRNA synthetases in Halobacterium salinarum, our model extremophile organism. Both P. intermediaand P. gingivalis are human pathogens that cause the diseases Noma and Cancrum Oris (a facial flesh eating disease). These diseases mostly afflict countries that do not have access to adequate dental and medical care. We have cloned and expressed the arginyl-tRNA synthetase and the cysteinyl-tRNA synthetase from P. intermedia into E. coli. The goals of our summer projects will be to further characterize these enzymes, using protein mutagensis, enzymology, gel shift assays, and CD/fluorescence spectroscopy. Our ultimate goal is to determine if their “extreme” features could be exploited for potential drug development.

Courtney Jenkins, Ph.D.
Assistant Professor
Office: Office: PSC 347, Pocatello
Summary: The Jenkins group focuses on creating sulfur-based polymers through a process called inverse vulcanization. Elemental sulfur is a waste product generated during the purification of petroleum. Sulfur can be combined with a variety of monomers including many renewable monomers in a solvent-free synthesis to form inexpensive polymers. These materials can be used for a variety of applications including as adhesives and heavy metal sorbents.
Projects: Each project will begin by synthesizing polysulfides. One project will focus on determining how the incorporation of different monomers impacts the solubility and swelling of these polymers. The ultimate goal would be to develop modifiable hydrogels to use to study bacterial growth. A second project will incorporate renewable monomers to develop biomimetic adhesives. Each project will involve studying the chemical and physical properties of these polymers to determine how factors such as the structure and polymer chain length impact the materials’ function

Joshua Pak
Department Chair - Professor
Office: PSC 145/360
Summary: Research interests in Pak Laboratory consist of the preparation and study of non-natural nano- to meso-scale materials. We develop and utilize modern synthetic methods for the preparation of novel materials with technologically important properties. Our core strategy is to design functional materials through organic and inorganic synthetic chemistry. Some of the target materials in our research projects include novel organic ligands, oligomers/polymers, organometallic compounds, metal chalcogenide nanoparticles/thin films and organic-inorganic hybrid materials.
Minimum Classes: Pak lab frequently employs high school student, high school teachers, undergraduate students of all levels. As an INBRE fellow, it is helpful to have completed Organic Chemistry and/or Inorganic Chemistry courses, but not required.
Preparation and study of organic and organic-inorganic hybrid materials for drug delivery and other applications
Metal-Organic Frameworks (MOFs) are highly crystalline porous organic-inorganic hybrid materials with extremely high surface areas. In recent years, MOFs have been studied for their potential utilities in gas separation, environmental remediation, and drug delivery applications to name a few. An INBRE fellow would be involved in syntheses of novel ligands and related MOFs along with investigation of fundamental behaviors of MOFs such as structure, stability, and their host-guest chemistry of potential drug molecules.
Preparation and study of polymers and oligomers for drug delivery and other applications
Polymers and oligomers (polymers with a very few repeat units) are important biomedical materials with variety of applications. Some polymers and oligomers have been used in drug delivery, such as cancer therapeutics and mRNA, some have been used as biocompatible coating materials in implants, and some have been used as medical adhesives or biodegradable suture. An INBRE fellow would be involved in syntheses and characterization of novel polymers and oligomers while examining their structure-property-relationships in terms of their physical and chemical properties under a variety of conditions.
.jpg)
Leslie Nickerson
Assistant Professor
Office: PSC 36
Summary: Research in the Nickerson lab is focused on adapting traditional organic transformations that use metal-halide based Lewis acids to more sustainable processes using Lewis acidic heterogeneous materials such as clays and zeolites. This adjustment makes methods safer and more sustainable because the heterogeneous materials are easily recycled and reused and the waste production is far less than when using traditional Lewis acids. Current projects are focused on modifying the Friedel-Crafts acylation, ester hydrolysis, and the Prins-pinacol rearrangement.
Minimum classes: Preference will be given to students that have completed organic chemistry I and II.
Projects: Summer students can be incorporated into either of the projects to help establish consistent and reliable methodologies. They can also be part of the substrate scope expansion to identify the limitations for each project. The proposed research will provide student researchers valuable skills in organic synthesis, method development and materials chemistry. Students will learn organic chemistry techniques, experimental design with an emphasis on safety considerations, data analysis, and spectroscopic techniques including NMR and GC. This work will be ideal for any students that want to learn more about implementing sustainable methods in organic chemistry.

Ali Habashi
Associate Professor
Office: Eames Complex
Summary: I am interested in exploring the concept of drug-disease interaction, and studying the effect of inflammation, in particular, on the renin-angiotensin system (RAS) at enzyme, peptide, and receptor levels in order to fully understand the underlying mechanisms. In different inflammatory conditions such as rheumatoid arthritis, cancer, diabetes, mental disorders, and Alzheimer disease, patient’s quality of life has been be affected by the deleterious impact of inflammation. Due to extensive involvement of the RAS in the systemic and local regulatory function of different organs and the significant impact of inflammation on the activation of the RAS, the association of the RAS in different pathological conditions has been reported. The RAS consists of two counteracting arms: tissue protective and tissue toxic. Manipulation of the RAS through augmentation of its tissue protective arm by delivering of its peptide homologs seems promising. Peptides have gained increased interest as biological therapeutics during recent years. However, the clinical application of these agents is still limited due to drug delivery challenges. As a pharmaceutical formulation scientist, I have set focuses of my lab on exploring innovative targeted drug delivery systems for effective, safe, and noninvasive delivery of these therapeutic agents for aiming at several serious inflammatory conditions that RAS involved in their pathology.
Minimum classes: It is beneficial for the student’s adaptation into the laboratory environment if he/she has any knowledge of introductory to science, biology and inorganic and organic chemistry, but it is not required.
Projects: Some of the major projects that an undergraduate student can get involved in my lab are synthesis, separation and characterization of PEGylated conjugates of small peptides using analytical chemistry and instrumental analysis methods and testing the biological activity of conjugates using cell culture and studying their absorption, distribution, metabolism and elimination in small animals. Experiments will be performed by graduate students under supervision of the principle investigator. These project provides opportunities for one or more undergraduate researchers to join the lab in order to be introduced into the biomedical and pharmaceutical research and learn different laboratory techniques such as instrumental analysis, sample preparation methods, cell cultures, western blotting, animal models of different disease, pharmacokinetics studies and so on. These skills are in high demand by Pharmaceutical and Biotech companies. Early exposure to such research environment will benefit an enthusiastic, energetic and motivated undergraduate student to be able to plan his/her career ahead of the other competitors for their cohort.

Danny Xu
Associate Professor, Director of BPSCI Graduate Programs and Admissions
Office: Meridian 752
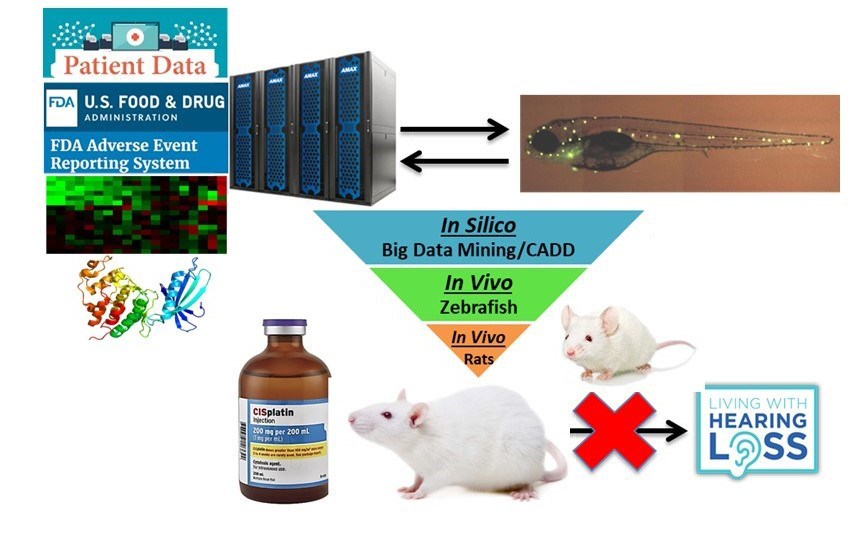
Summary: Hearing loss is a significant public health issue, affecting 38 million Americans and 500 million people worldwide. Dr. Xu’s lab is working to identify novel therapeutics for hearing loss prevention utilizing an array of state-of-the-art discovery tools, including big data mining, molecular modeling, fluorescence confocal imaging, zebrafish, and mammalian animal models.
Projects:
- Project 1: To identify new ototoxic drugs and drug combinations
- Project 2: To identify new otoprotective drugs and drug combinations

Jesse Jones
Assistant Professor
Office: Eames Complex
Summary: My research interests revolve around the development of targeted therapeutics and include an interdisciplinary approach spanning biological chemistry, drug discovery, molecular pharmaceutics, and synthetic biology. My current focus employs a multi-faceted approach comprised of classical drug discovery efforts that include the characterization and validation of specific enzyme- and structural-protein targets for the development of selective antimicrobials; as well as more advanced synthetic biology efforts including discovery, characterization, and advanced engineering of protein nanocages for the development of targeted drug and therapeutic prodrug nanoreactor delivery platforms.
Projects:
1. Biomedical engineering and synthetic biology: development of therapeutic prodrug nanoreactors and drug delivery platforms. This project relies on the tractable nature of protein nanocages and assemblies, as well as the selective nature of protein-protein interactions (such as antibodies and antibody mimetics) that allow for targeted delivery of therapeutic proteins. This project includes implementing cutting-edge cloning and molecular biology techniques, such as optimized protein-protein interactions, “molecular superglues,” and “click-chemistry,” to rationally manipulate tractable microbial protein nanocage scaffolds such as bacterial encapsulins and viral capsids. This includes internal engineering for rational drug, enzyme, and gene cargo loading, as well as external engineering for anti-immunogenic and targeting moieties to develop sophisticated therapeutic payloads capable of focused delivery to specific proteins, cells, and organs of interest.
2. Classic drug discovery: selective enzyme and structural protein drug target characterization, validation, and development. This project hinges on the fact that certain enzymes, isozymes, and structural proteins are uniquely essential to certain pathogenic microbes—but not to commensal or beneficial microbes—and therefore represent promising targets for eliciting selective antimicrobial effects against only the targeted pathogens of interest. This project includes classic cloning, bioinformatics, protein biochemistry, assay development, drug screening (high throughput and in silico), and structural biology techniques. Techniques learned and applied are likely to include bioinformatics, polymerase chain reaction (PCR), fast protein liquid chromatography (FPLC), plate reader absorbance- and fluorescence-based high-throughput screening (HTS), in silico computational chemistry, in silico automated protein structure prediction, negative-stain transmission electron microscopy (TEM), and more.

Kumari Kavita Sharma
Assistant Research Professor
Office: Eames Complex
Summary: My research expertise is in chromatographic and spectroscopic techniques. The lab work comprises development and validation of the analytical methodologies with the use of highly sophisticated techniques/instruments for the identification, characterization and quantification of the secondary metabolites/bio marker compounds in a wide range of matrices. The samples come from natural products, biological materials, microorganisms, environment, ionic liquids and synthetic compounds. So, the gamut of my entire work incorporates mass spectrometric related analytical techniques. Some of the equipment that we use for resolving the research challenges are high performance liquid chromatography (HPLC), preparative HPLC (Prep-HPLC), gas chromatography mass spectrometry (GC-MS), liquid chromatography mass spectrometry (LC-MS), tandem mass spectrometry (LC-MS/MS), surface plasmon resonance (SPR), nuclear magnetic resonance (NMR) spectroscopy and UV/Vis spectrophotometer.
Minimum Classes: N/A
Projects:
- Extraction and characterization of natural products in the leaves of Artemisia tridentate: The leaf foliage is extracted with 100 % chloroform. The extracts are then characterized by thin layer chromatography (TLC), high performance liquid chromatography (HPLC), and extensive spectroscopic analysis, including 1D- and 2D-nuclear magnetic resonance (NMR), Fourier transform infrared (FTIR) spectroscopy, and mass spectrometry (MS). In vitro and in vivo studies will be done on the compounds isolated from the extracts to check their potency for antioxidant activity and anticancer properties. In addition, derivatization of the most abundant compound will be carried out to understand the Structure-Activity Relationship (SAR) of the modified compounds. Isolated compounds and their derivatives will be used for the phenotypic studies in the zebrafish.
- Metabolomics of the most dominating gut microbes for the treatment of inflammatory bowel disease (IBD), type 2 diabetes, neurological disorders, cardiovascular disease, respiratory illness and cancers: Gut microbiota is a large reservoir of microorganisms, including bacteria, archaea, fungi and viruses living in the digestive tract of animals. It makes up more cells than all the rest of our body put together. It has specific functions, maintains our gut health, and controls the remote extra enteric organs such as the liver, brain, skin, heart, muscle, and bone via bidirectional signaling process. They interfere with various pathways involved in immunity, energy, and lipid and glucose metabolism by releasing metabolites, enzymes and hormones. The gut derived metabolite serves as a potential biomarker for the early diagnosis of metabolic disorders. We will be analyzing the composition of the metabolites derived from the gut microbiomes to get a better understanding of their roles and functionality in disease development, progression and immune modulation.

Sarah Hobdey
Research Assistant Professor
Office: Meridian 761 Boise VAMC 203
Summary: Research in my laboratory is centered on the development of novel therapies for the treatment of life-threating necrotizing soft tissue infections caused by Gram positive bacteria. Morbidity and mortality from infections caused by Lancefield groups A, C and G streptococcus, Staphylococcus aureus including methicillin-resistant species (MRSA), and multiple clostridial species remains exceedingly high despite modern antibiotic regimens and intensive care measures. Through the production of potent, well-characterized, extracellular toxins, each organism causes rapidly progressive necrotizing infections that destroy skin, subcutaneous tissue, fascia and muscle, often leading to multiple organ failure, toxic shock syndromes and death. Survivors often require surgery, including amputation, necessitating prolonged hospitalization and rehabilitation. The most important factor contributing to poor patient outcomes is a delay in treatment, which permits bacterial proliferation and toxin production to proceed unchecked. To improve survival and reduce morbidity, we are pursuing a passive immunization strategy that can be delivered to the patient immediately upon clinical presentation and will neutralize the damaging toxins as they are secreted by the suspected pathogen(s).
Minimum classes: None
Projects: Our research relies on the isolation of B-cells that produce toxin-specific antibodies, cloning of these antibody genes and generating recombinant forms of these antibodies in vitro. In order to achieve this we must to produce recombinant bacterial toxins as ‘bait’ to isolate the toxin-specific B-cells, Project 1. Once the toxin-specific B-cells have been isolated and cloned, we need reproduce the human monoclonal antibodies in vitro using a mammalian expression system and characterized their ability to neutralize the specific bacterial toxins, in vitro and in vivo, Project 2.
Project 1) Creation of recombinant bacterial toxins used for isolation of toxin-specific B-cells. Techniques may include: bacterial expression, molecular cloning, polymerase chain reaction, protein purification utilizing fast protein liquid chromatography (FPLC), in vitro biochemical protein characterizations, spectrometry, flow-cytometry, polyacrylamide gel electrophoresis (PAGE), western blot.
Project 2) Development and characterization of recombinant anti-toxin antibodies. Techniques may include: cell culture, restriction enzyme-free cloning, transfection, site-directed mutagenesis, protein purification utilizing FPLC, in vitro (toxin neutralization assays), ex vivo (murine cardiomyocyte dysfunction) or in vivo (murine models of necrotizing infection) characterization of recombinant antibodies.

Solomon Zeleke
Assistant Professor
Summary: My medicinal chemistry research program investigates the discovery and development of small-molecule inhibitors targeting cyclin-dependent kinases (CDKs). I have identified novel clinical stage CDK4-, CDK4/6-, and CDK12-selective inhibitors as candidate therapeutics for cancer. I am interested in the design and synthesis of CDK-selective inhibitors, and my current efforts are focused on discovering novel CDK-selective inhibitors and degraders.

Srinath Pashikanti
Assistant Professor
Office: Eames Complex
Summary: Our group utilizes organic chemistry towards synthesis of cell permeable medicinally active small molecules. For example, we are developing small molecules in understanding the physiological and biochemical role of ceramide metabolizing enzymes. Ceramide is a bioactive sphingolipid that exhibits anticancer properties in a cell. Strategies aimed at increasing the cellular ceramide induce apoptosis in cancer cells. To complement our synthetic efforts, we perform in vitro experiments and cell based assays towards determining the biological activity of these analogs in a structure-activity relationship model.
Minimum classes: General chemistry/organic chemistry/biochemistry
Projects: A. Synthesis, structure-activity relationship studies of small molecules in understanding the biological relevance of ceramide metabolizing enzymes. This project involves synthesis of natural product based analogs or natural product-like analogs followed by in vitro based assays towards quantification of cellular ceramide levels. Several studies demonstrated that increase in cellular ceramide concentration resulted in inducing apoptosis in cancer cells. We hypothesize that by blocking ceramide metabolism we can modulate the cellular ceramide concentrations and induce apoptosis in cancer cells.
B.Identification of medicinally active small molecules to inhibit glycation of histone proteins. Glycation, a type of covalent modification of protein, observed at higher incidence under oxidative stress conditions. Histones maintain DNA integrity. Glycation of histones at the genomic level disrupts the integrity of DNA resulting in deleterious effects. We hypothesize that by minimizing these covalent modifications of histones especially under oxidative stress conditions we can minimize the DNA damage. We use natural product based phytochemicals as glycation inhibitors.
Our research projects outlined above are collaborative and multidisciplinary in nature. Students trained in biochemistry or organic chemistry or both disciplines can participate in these projects.

Erin Rasmussen
Professor
Office: Garrison Rm 411
Summary:
Minimum Classes:
Projects:

Maria Wong
Professor
Office: Garrison Rm 418
Summary:
Minimum Classes:
Projects:

Michele Brumely
Professor, Experimental Psychology - Associate Vice President for Research
Summary: My developmental behavioral neuroscience lab studies the development of motor behavior, and the role that neural systems and experience play in shaping coordinated behavior. Of particular interest is the development of locomotion, posture, reflexes, the spinal cord, and epigenetic changes in the central nervous system. Ongoing studies are studying these issues in rats.
Projects: INBRE fellows may be involved in collecting and analyzing data for behavioral neuroscience research experiments with developing rats. Types of projects include investigating locomotor and motor reflex behavior in the newborn rat, and studying maternal-infant interactions in rats. Techniques used may include different types of drug administration, spinal cord surgeries, microscopy, different methods for manipulating sensory feedback and testing motor behavior, behavior recording, and behavior scoring.