
Dr. John Kalivas
Professor Emeritus
Office: PSC 253A, Pocatello
208-282-4373
Ph.D. Analytical Chemistry, University of Washington, 1982
Research areas: Analytical Chemistry, Chemometrics, Chemical Education
Student experience required for research: CHEM 1111 and CHEM 1112
Student experience gained from research: Data analysis, modeling, computational chemistry, spectroscopy, teaching
Research Focus
Our analytical chemistry research focus is in an area termed chemometrics concerned with “how to obtain chemically relevant information out of measured chemical data, how to represent and display this information, and how to get such information into data.”1 While this area of research was well established in the early 1970’s, the area is now popularized as data science, machine learning (ML), and artificial intelligence (AI).
We concentrate on developing computer algorithms to determine mathematical relationships between chemical data and analyte properties desired such as the concentration estimate of a substance in a sample. Algorithms are trained to recognize data patterns and predict properties for new situations. For example, training an algorithm to distinguish noninvasively measured spectra of cancer cells from non-cancer cells. Our mission is creating new algorithms to solve difficult analytical chemistry problems by leveraging sample and measurement matrix effects (hidden physicochemical properties) as information to work with. Our current emphasis is using the immersive analytics tool virtual reality (VR) allowing the human to see, touch, and hear the data in order to improve on data decisions compared to autonomous algorithms including our own. Some research projects follow.
Multivariate Calibration (Modeling): Central to many disciplines is multivariate calibration including food analysis, food adulteration detection and authentication of product origin, environmental monitoring, industrial process control, medical diagnosis
such as disease detection, pharmaceutical analysis, forensic analysis, detection of hidden radioactive material, and the list goes on. Work in our laboratory consists of developing new mathematical processes and the corresponding computer algorithm implementations in order to improve calibration quality in conjunction with eliminating user decisions making calibration and subsequent analyte predictions automatic. A persistent multidisciplinary problem is developing a calibration model in one set of environmental, instrumental, physicochemical conditions (the primary source conditions) to now work in new target conditions, i.e., the calibration maintenance (transfer) problem. In ML language, calibration is called training and unknown samples to be predicted are named the target samples.
Our laboratory developed two approaches to solve this problem that now allow on-site analyses with handheld devices including consumer applications. One solution is local modeling.
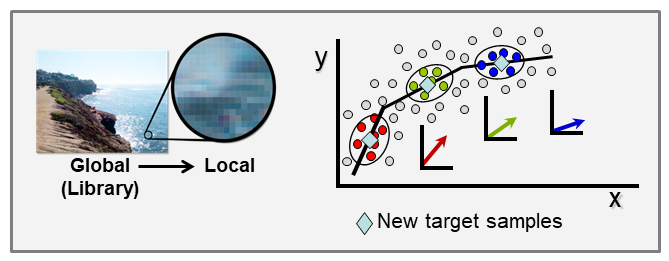
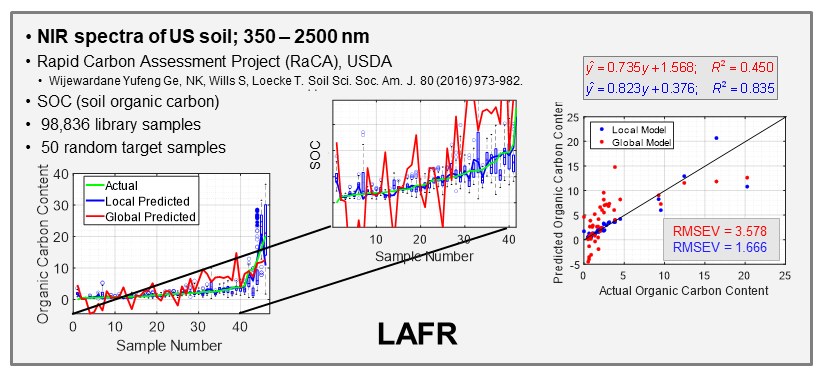
With the ever-growing availability of spectral data, e.g., near infrared (NIR), Raman, fluorescence, and other spectral processes, there is a high need for computer algorithms to mine through such data libraries and identify those spectra similar to a spectrum just measured for a new target sample. The user is interested in using the new spectrum for quantitative analysis of an analyte, perhaps glucose content for diabetic, and a model must first be formed using calibration spectra matrix matched to the new target sample spectrum. Because the analyte amount in the new sample is not known, identifying fully matched calibration samples from a library is not a simple task. However, we have developed an algorithm to accomplish this objective that we call local adaptive fusion regression (LAFR). Key takeaways for LAFR are: (1) models are formed by an understandable process based on physicochemical properties and principles with a linear Beer’s law-like relationship, (2) final calibration sets bracket target sample analyte values, and (3) all adjustable parameters are self-optimized. Ongoing work involves providing a reliability measure based on the probability of a correct prediction as well as using VR to further refine the LAFR calibration set to those samples best matched to the target sample.
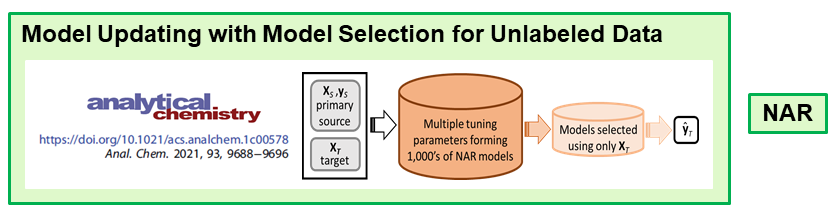
The second approach is model updating by the transfer learning approach domain adaptation. In collaboration with Erik Andries at Central New Mexico Community College, we now have a null augmented regression (NAR) algorithm that correctly predicts new target samples. Models are formed using the new unlabeled target samples for which is prediction is desired.
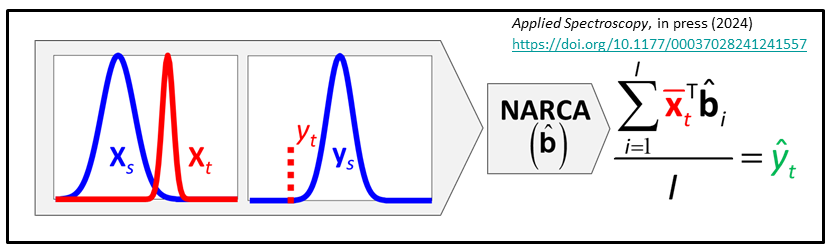
 We have just developed NAR constant analyte (NARCA) as a ne local modeling approach where the multiple spectra are measured from a single sample and eventually transformed to the analyte amount present in the target sample. The sample can be homogeneous or slightly heterogeneous.
We have just developed NAR constant analyte (NARCA) as a ne local modeling approach where the multiple spectra are measured from a single sample and eventually transformed to the analyte amount present in the target sample. The sample can be homogeneous or slightly heterogeneous.
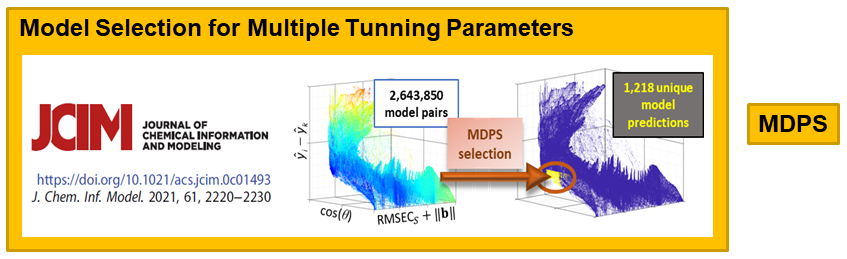
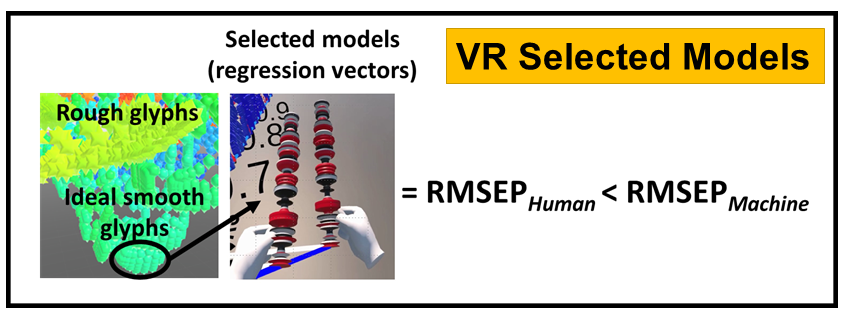
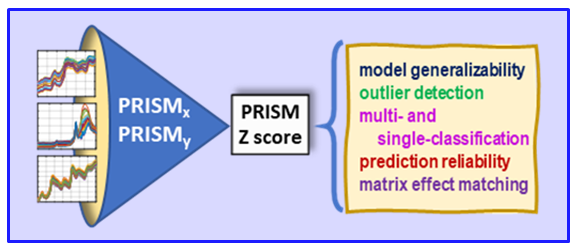
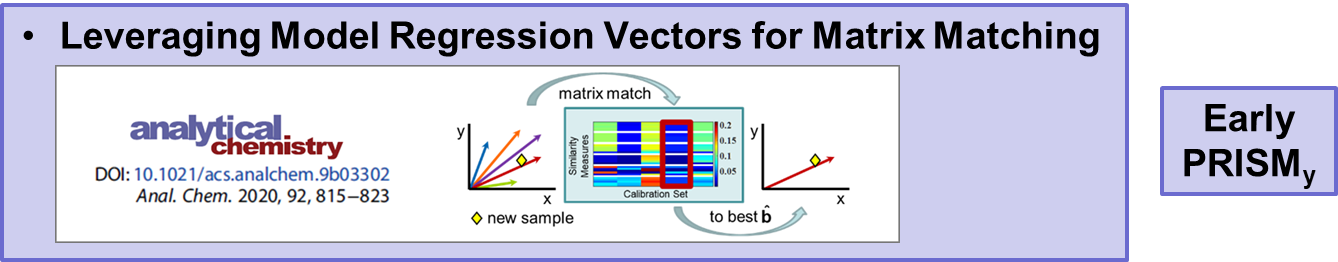
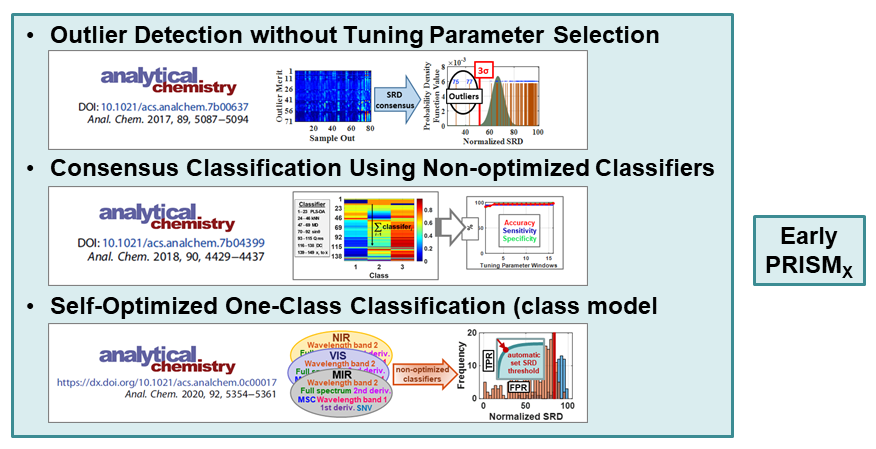
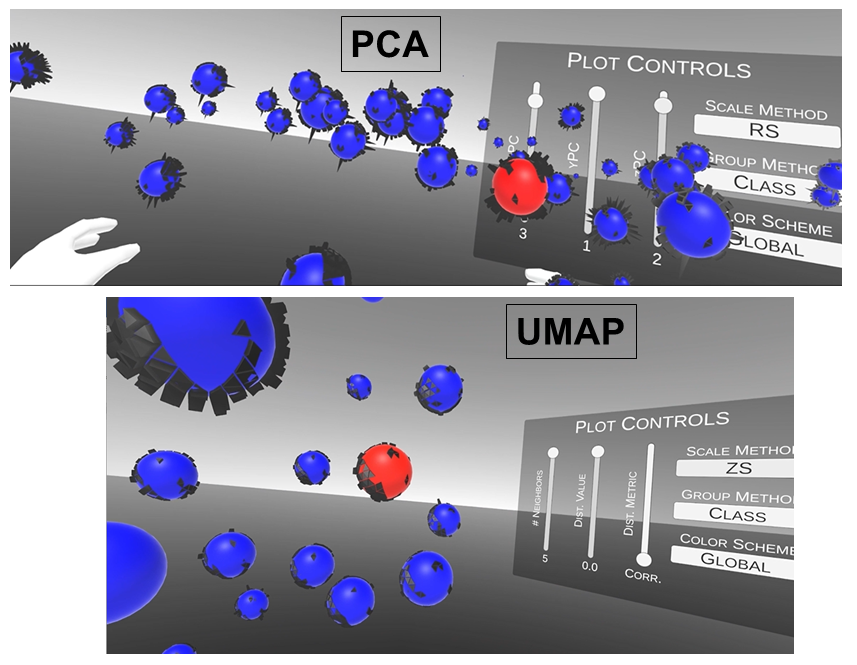
VR and Vision Impaired: As noted above, we have various ongoing VR projects. The goal is to develop hybrid systems relying on the computer for the intense chemometric calculations and VR for the final data analysis decisions. By using VR, samples have spatial relationships as with any flat screen graphic but with PRISM rendered in VR, we are able to provide a shape to each sample data point. Shapes are relative to the sample-wise matrix effects determined by PRISM.
Ultimately, we are going to use our VR data analysis for the vision impaired where the user will be able to hear where data clouds are and then using haptic gloves feel the difference between the samples for deciding if a sample is an outlier, or a class member, or which calibration samples are matrix matched to the target sample for local modeling.
- Wold. “Chemometrics; what do we mean with it, and what do we want from it?”, Chemometorcs and Intelligent Laboratory Systems, 30 (1995) 109-115.
Recent Publications
H.J. Redd, J.H. Kalivas: “Assessment of Conformal Prediction and Standard Normal Distribution for Autonomous Consensus One-Class Classification”, Journal of Chemometrics, in press (2025) https://doi.org/10.1002/cem.3639
R.C. Spiers, J.H. Kalivas: “Local Adaptive Fusion Regression (LAFR) for Local Linear Multivariate Calibration: Application to Large Datasets”, Applied Spectroscopy, in press (2025) https://doi.org/10.1177/00037028241308538
J.M.J. Peper, J.H. Kalivas: “Redefining Spectral Data Analysis with Immersive Analytics: Exploring Domain Shifted Model Spaces for Optimal Model Selection”, Applied Spectroscopy, in press (2024). https://doi.org/10.1177/00037028241280669
J.M.J. Peper, J.H. Kalivas: “Local Modeling by Adapting Source Calibration Models to Analyte Shifted Target Domain Samples without Reference Values”, Applied Spectroscopy, in press (2024) https://doi.org/10.1177/00037028241241557
R. Spiers, C. Norby, J.H. Kalivas: “Physicochemical Responsive Integrated Similarity Measure (PRISM) for a Comprehensive Quantitative Perspective of Sample Similarity Dynamically Assessed with NIR Spectra”, Analytical Chemistry, 95, 12776–12784 (2023) https://doi.org/10.1021/acs.analchem.3c01616
J.H. Kalivas, T. Lemos: “Automatic Food and Beverage Authentication and Adulteration Detection by Classification Hybrid Fusion”, Journal of Chemometrics, 37(3) (2023):e3371 https://doi.org/10.1002/cem.3371
MATLAB Code
For MATLAB code to preform Tikhonov regularization without reference samples, see MATLAB Code
For MATLAB code to preform sum of ranking differences (SRD), see 2013_12_16_SRD.7z
For MATLAB code to preform fusion classification, see 2018_3_1_ClassificationCode.7z
For MATLAB code to preform Model updating by sample and feature augmentation, see 2018_9_13_SAFA
For MATLAB code to preform single-class fusion classification with SRD, see 2019_11_25_SingleClassCode.7z
For MATLAB code to preform model selection by model diversity and prediction similarity, see 2020_7_1_MDPS.7z
For MATLAB code to perform physicochemical responsive integrated similarity measure (PRISM) with SRD, see PRISM_SRD.7z
For MATLAB code to perform null augmented regression constant analyte (NARCA), see NARCA.7z

